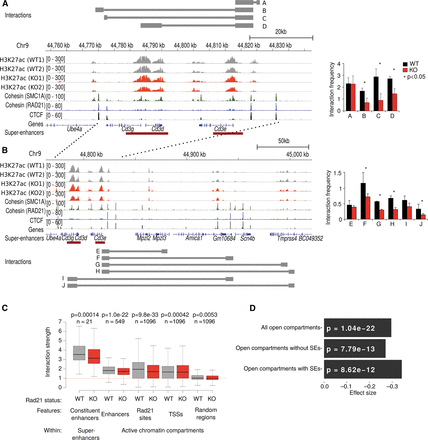
Cohesin mediates the spatial clustering of enhancer elements. (A) The mouse Cd3 super-enhancer is flanked and punctuated by CTCF and cohesin binding (refer to Fig. 2A,B for additional examples of the relationship between super-enhancers, CTCF, and cohesin). Restriction fragments used for 3C analysis are indicated by gray bars. “A” is a proximity ligation control that demonstrates comparable efficiency of 3C experiments. Enhancer-enhancer interactions B, C, and D across the Cd3 super-enhancer were significantly reduced in Rad21-deficient (red) compared to control (black) thymocytes. n = 3, mean ± SD, (*) P < 0.05. (B) Long-range interactions between the Cd3 super-enhancer and enhancer elements outside the Cd3 locus. “E” is a control used to demonstrate background interactions with a downstream genomic fragment lacking H3K27ac marks and CTCF binding. The position of the Cd3 super-enhancer is marked by dashed lines. Interactions F, G, H, I, and J link the Cd3 super-enhancer with downstream enhancer elements outside the Cd3 region and were significantly reduced in Rad21-deficient (red) compared to control (black) thymocytes. n = 3, mean ± SD, (*) P < 0.05. (C) Structured interaction matrix analysis (SIMA) of long-range interactions between chromatin features based on Hi-C data for control (gray) and Rad21-deficient (red) thymocytes. Interactions between constituent elements of super-enhancers (left) were analyzed by SIMA within super-enhancer regions sized 100 kb and larger. Interactions between other chromatin features were analyzed by SIMA within open chromatin compartments. Note that chromatin features show increased self-interactions, while interactions of random regions (Seitan et al. 2013) conformed to the level of interactions predicted by a background model based on genomic distance and sequencing depth (dashed red line). “Interaction strength” refers to the strength of interactions between 10-kb regions within super-enhancers (SEs) or 10-kb regions within chromosomal compartments (Enh, RAD21 sites, TSS, and random regions after normalization to the background model). P-values shown are from a Wilcoxon signed-rank test. (D) Reduced enhancer-enhancer interactions in Rad21-deficient thymocytes based on SIMA analysis of Hi-C data are compared for all open chromosomal compartments and separately for compartments with or without super-enhancers. P-values shown are from a Wilcoxon signed-rank test.











