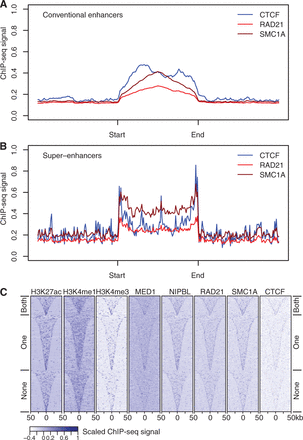
Enhancer elements are enriched for CTCF and cohesin binding. (A) CTCF, RAD21, and SMC1A ChIP-seq signal enrichment in thymocytes at conventional enhancers. Enhancers were defined based on H2K27ac ChIP-seq data (Methods) and enhancer length was normalized in order to align the start and end of the enhancer. (B) CTCF, RAD21, and SMC1A ChIP-seq signal enrichment in thymocytes at super-enhancers. Super-enhancers were defined based on H2K27ac ChIP-seq data (Methods), enhancer length was normalized in order to align the start and end of the super-enhancers, and flanking regions of equal size to the super-enhancer are shown for reference. “Start” and “end” are based on the genomic coordinates. (C) Classification of super-enhancers on the basis of CTCF binding at both boundaries, one boundary, or neither boundary. Heatmaps of ChIP-seq signal enrichment in 100-kb windows around super-enhancer centers, grouped according to CTCF binding at the super-enhancer boundaries. Histone modifications, the cohesin loading factor NIPBL, and the cohesin subunits RAD21 and SMC1A are shown for reference.











