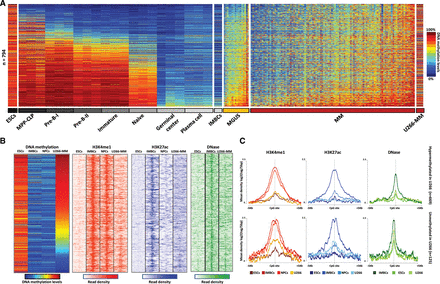
DNA methylation and chromatin features of hypermethylated enhancers in MM in the context of normal B-cell differentiation. (A) DNA methylation levels of 794 enhancer-associated CpGs in seven B-cell differentiation stages, MGUS and MM patient samples, as well as ESCs (H1), IMBCs (GM12878), and U266-MM cell lines. (B) ChIP-seq levels of H3K4me1 and H3K27ac, and DNase-seq data of 794 enhancer-associated CpGs in ESCs, IMBCs, NPCs, and the U266 MM cell line. (C) Density plot of H3K4me1, H3K27ac, and DNase levels in ESCs, IMBCs, NPCs, and the U266 MM cell line. Among the 794 enhancer-associated CpGs in MM, those hypermethylated in U266 are shown in the top panel, whereas those unmethylated in this cell line appear at the bottom. (MPP-CLP) Hematopoietic multipotent progenitors–common lymphoid progenitors; (ESCs) embryonic stem cells; (IMBCs) immortalized mature B cells; (NPCs) normal plasma cells; (U266-MM) multiple myeloma derived cell line U266.











