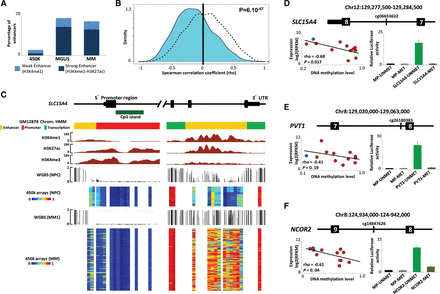
Functional and transcriptional analysis of hypermethylated enhancer regions in MM. (A) Percentage of hypermethylated CpGs associated with enhancer regions in MGUS and MM patient samples using HumanMethylation450 BeadChip data. (B) Density plot of correlation coefficients between methylation levels of hypermethylated enhancers and the expression of their associated genes (cyan) using RNA-seq data. As control, intronic CpGs of the same genes were studied excluding the enhancer associated ones (black dotted line). For this analysis, we used 663 CpGs (out of 794) annotated to 574 genes with available gene expression, and 8956 CpGs in nonenhancer intronic regions of the same genes. (C) A snapshot of the UCSC Genome Browser showing the promoter (left) and the 3′ intronic enhancer region (right) of the SLC15A4 gene. Displayed tracks include the chromatin state characterization in IMBCs and ChIP-seq data for H3K27ac, H3K4me1, and H3K4me3. DNA methylation levels of NPCs and MM patient samples measured by WGBS and HumanMethylation450 BeadChip are also shown. (D–F) Correlation analysis between DNA methylation levels of the hypermethylated enhancers and expression of the associated gene (left). In addition, we display the luciferase reporter activity data of the analyzed enhancer region (right) located in the intron of (D) SLC15A4, (E) PVT1, and (F) NCOR2. (MP) Minimal promoter; (MET) methylated; (UNMET) unmethylated.











