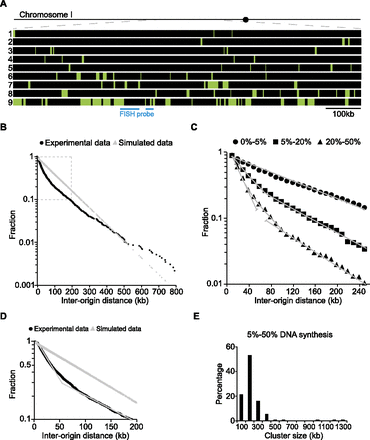
Replication origins both fire stochastically and form clusters. (A) Replication profile of nine completely overlapping 1-Mb single DNA molecules corresponding to the same region on Chromosome I (from 2100 to 3100 kb). (B) Semilog plot of the cumulative frequency of IODs measured for 131 single DNA molecules with an average length of 1.8 Mb and replicated up to 50% (black curve). The values on the y-axis correspond to the fraction of IODs that are larger in size than the corresponding IOD on the x-axis. The gray line corresponds to simulated data for a purely random distribution of the same number of origins over the same DNA length. The experimental measurements deviate significantly from the purely stochastic prediction, P < 0.001 (Lilliefors statistical test). (C) Semilog plot of the cumulative frequencies of IODs for molecules sampled according to their extent of DNA synthesis; black circles, black squares, and black triangles correspond to molecules replicated from 0%–5%, 5%–20%, and 20%–50%, respectively. At the beginning of S-phase, origin selection is random, as the cumulative frequency of IODs for molecules replicated up to 5% fits a straight line. For molecules replicated from 5%–20%, the curve deviates from the straight line at short IODs. The curve corresponding to molecules replicated from 20% to 50% can be decomposed into two straight lines with different slopes. (D) Semilog plot of the data within the dashed box in B, representing 90% of the IODs. The experimental data curve can be decomposed into two straight lines with different slopes (thin gray line), indicative of two regimes of stochastic origin selection operating along chromosomes. (E) Histogram of cluster sizes for molecules replicated from 5% to 50%.











