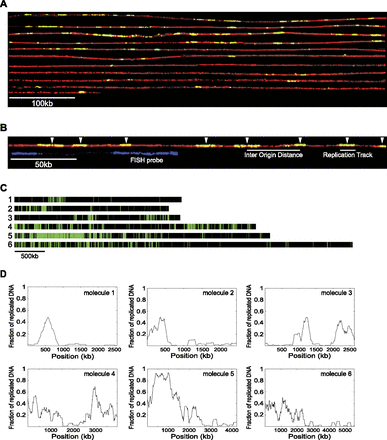
Visualizing DNA replication on single DNA molecules using DNA combing. (A) A single 5.6-Mb molecule that approximately spans the entire length of Chromosome I. To visualize individual BrdU tracks, the molecule was “cut” in silico into 21 consecutive fragments of 270 kb each, and a composite image was constructed. Newly synthesized DNA was labeled in vivo using the halogenated thymidine analog BrdU. Protein-free DNA was combed onto a silanized glass surface and visualized with anti-thymidine antibody (red), and incorporated BrdU was visualized with anti-BrdU antibody (green). BrdU tracks (replication tracks) shown in yellow (green overlaying red) correspond to newly synthesized DNA. (B) White arrows point to the middle of replication tracks (shown with a short white line below the DNA molecule) where we assume replication origins are located. The distance between the middle of two adjacent replication tracks represents the inter-origin distance (IOD) (shown with a longer white line below the DNA molecule). To detect and orient specific DNA sequences on combed DNA molecules, we used fluorescent in situ hybridization (FISH). Each chromosomal position is detected by a unique signature of two probes with differing lengths and distances between them (shown in blue below the DNA molecule), which allowed us to orient the combed DNA molecules and to align them to the corresponding chromosomal region. (C) The distribution of replication tracks on six single DNA molecules randomly selected from the population of molecules. Green bars represent BrdU replication tracks and black bars represent unreplicated segments on DNA molecules. Black and green bars are drawn to scale. All DNA molecules analyzed in this study are represented in Supplemental Figure S15, and the measurements for each molecule are listed in Supplemental Table S2. The DNA molecule shown in A corresponds to molecule 6 in C. (D) Moving average analysis using a 200-kb window size in 2-kb steps (the resolution of the DNA combing technique) of the six molecules shown in C. Each value corresponds to the fraction of replicated DNA within a 200-kb stretch of DNA. Peaks represent chromosomal domains rich in replicated DNA, whereas troughs represent chromosomal domains poor in replicated DNA.











