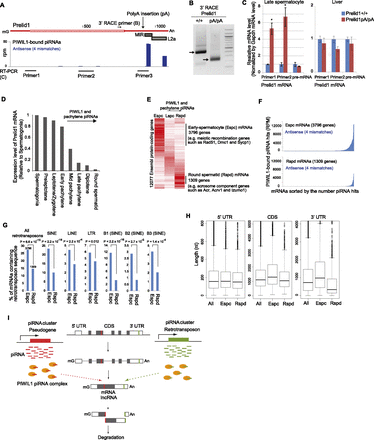
The mammalian conserved retrotransposon sequence in Prelid1 mRNA regulates its stability in late spermatocytes. (A) The distribution of PIWIL1-associated piRNAs mapped to Prelid1 mRNA in the antisense orientation. PIWIL1-associated piRNAs are mapped to the MIR and L2 retrotransposon sequences (gray boxes) in the antisense orientation. (B) 3′RACE analysis of Prelid1 mRNA in Prelid1pA/pA late spermatocytes. The arrows indicate 3′RACE PCR products amplified from the 3′ ends of wild-type and truncated Prelid1 mRNAs. The insertion site of the polyadenylation signal sequence and the location of the primer used are shown in A. (C) q-PCR analysis of Prelid1 mRNA and pre-mRNA in Prelid1pA/pA late spermatocytes (left) and livers (right). Error bars represent the SE (n = 3). (*) P < 0.05 (Student’s t-test). The locations of the primers used for the analyses of the mRNA are shown in A. (D) Published microarray data (1431420_a_at probe) are shown (Fallahi et al. 2010). (E) Clustering analysis of the relative expression levels of 12,077 Ensembl protein-coding genes among early spermatocytes (Espc), late spermatocytes (Lspc), and round spermatids (RS) is shown. Expression levels were determined by RNA-seq in individual cell types. (F) The read number (RPM) of PIWIL1-associated piRNAs complementary to mRNA(s) of each gene is shown. (G) Retrotransposons are less frequently found in round spermatid (RS) mRNAs than early spermatocyte (Espc) mRNAs. The number of genes in each class is shown above the bar on the left graph. P-values between the <1 and >2 groups were calculated using the R function prop.test. (H) RS mRNAs have a short 3′ UTR. Box plots analyzing the length of 5′ UTR, CDS, and 3′ UTR of all Ensembl mRNAs, Espc mRNAs, and RS mRNAs are shown. (I) PIWIL1 and piRNAs derived from pseudogenes and retrotransposons in piRNA clusters degrade spermatocyte-expressed mRNAs and lncRNAs at the mature RNA level via piRNA-complementary sequences in the RNAs. The PIWIL1 and piRNA-induced degradation is mostly dependent on the slicer activity of PIWIL1.











