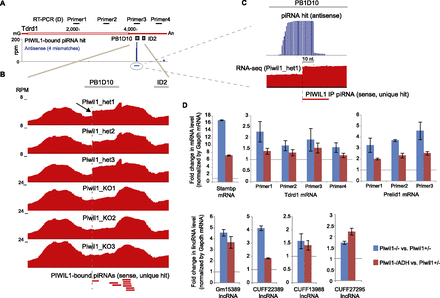
PIWIL1 slicer activity contributes to the degradation of target RNAs. (A) PIWIL1-associated piRNAs are predominantly mapped to PB1D10 SINE sequences (gray box) in the 3′ UTR of Tdrd1 mRNA. (B) The sharp increase in RNA-seq reads at the PB1D10 region. Shown are RNA-seq reads and PIWIL1-associated piRNAs derived from this region (below). (C) A magnified view of the site of the sharp increase. The location is indicated by the blue line in A. (D) q-PCR analysis of several target mRNAs (top) and lncRNAs (bottom) in Piwil1 KO (blue) and in Piwil1 slicer mutant (red) mice. The locations of PCR amplicons are shown in Figures 3A (Stambp), 5A (Tdrd1), and 7A (Prelid1). Error bars represent the SE (n = 3).











