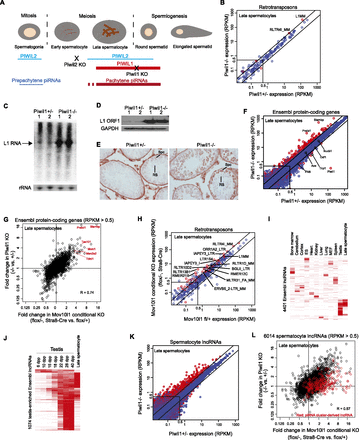
piRNAs negatively regulate mRNAs in spermatocytes. (A) A schematic drawing of mouse spermatogenesis, with the expression patterns of PIWIL1, PIWIL2, prepachytene piRNAs, and pachytene piRNAs. The stages of developmental arrest in Piwil1−/− and Piwil2−/− mice are shown. (B,F,K) RNA-seq comparison of retrotransposon (B), mRNA (F), and spermatocyte lncRNA (K) expression between Piwil1+/− and Piwil1−/− late spermatocytes. Lines flanking the diagonal represent twofold differences. Significantly changed genes (P < 0.025, Student’s t-test) are indicated in red (n = 3). (RPKM) Reads per kilobase of transcript per million mapped reads. (C,D) Northern blot analysis of L1 full-length RNA (C) and Western blot analysis of L1 ORF1 (D) in 23-dpp testes of Piwil1+/− and Piwil1−/− mice. Two biological replicates from different mice were analyzed. (E) Immunohistochemical analysis of L1 ORF1 in sections of 28-dpp testes from Piwil1+/− and Piwil1−/− mice. Spermatocytes (Spc) and round spermatids (RS) are indicated. (G,L) Piwil1 and Mov10l1 mutations have similar impacts on mRNA (G) and spermatocyte lncRNA (L) expression. lncRNAs derived from the top 50 pachytene piRNA clusters are indicated in red. (H) RNA-seq analysis of retrotransposon expression in late spermatocytes from Mov10l1 CKO and control mice. See B for more information. (I) Approximately 25% of lncRNAs registered in Ensembl are preferentially expressed in the testis. Shown are the expression levels of 4401 Ensembl lncRNAs (rows) across tissues and cells (columns). (J) Expression levels of 1074 testis-enriched Ensembl lncRNAs during spermatogenesis.











