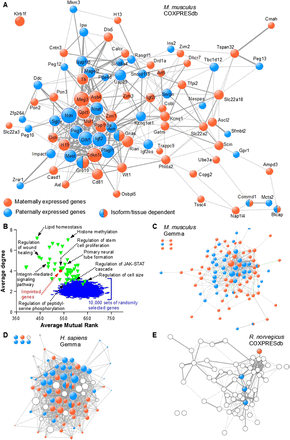
Imprinted genes are frequently coexpressed. (A) The COXPRESdb meta-analysis of microarray data was searched for coexpression among murine IGs. The resulting coexpression links are represented using Cytoscape. Node size is proportional to node degree. Edge width represents the mutual rank (see Supplemental Information) between two given nodes. (B) The average degree and average mutual rank were computed for the network of 85 IGs displayed in A (red circle). Random networks (blue lines) were generated by randomly drawing 10,000 sets of 85 Gene IDs with data in COXPRESdb and retrieving coexpression links as for the 85 murine IGs. A similar procedure was performed with 50 sets of 85 genes found in GO Biological Processes (green triangles) and with data in COXPRESdb (for a full list of the GO BPs analyzed, see Supplemental Table S22). (C) The Gemma meta-analysis of microarray data was searched for coexpression among murine IGs. Node size is proportional to node degree. Edge width maps the number of data sets in which two given nodes are coexpressed. (D) The Gemma database was searched for coexpression among human IGs and orthologs of murine IGs whose imprinting status in humans is unknown or uncertain (open circles) as in C. (E) The COXPRESdb was searched for coexpression of rat IGs and orthologs of murine IGs whose imprinting status in rat is unknown (open circles) as in A.











