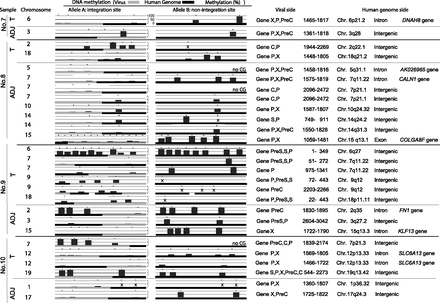
Allele-specific methylation analysis of the tumor (T) and adjacent nontumor (ADJ) sample genomes. The methylation levels of the HBV and human genomes for the integrated and unintegrated alleles in four paired tumor and adjacent nontumor samples (sample nos. 7–10) are shown. Detailed results of the HBV integrants (PreC, Precore; C, Core; PreS, Presurface; S, Surface; X, X) and flanking host genomes (position, chromosome, location of the genome, and gene names) are shown. The HBV genome became significantly methylated when integrated into highly methylated human genome regions, but not when integrated into unmethylated human genome regions. (×) The desired quantitative methylation levels were not obtained because of technical difficulties with the sequences that were being analyzed.











