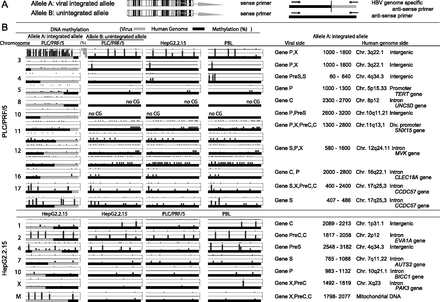
Allele-specific methylation analysis of the PLC/PRF/5 genome and the HepG2.2.15 genome. (A) A schema of allele-specific methylation analysis. (B) The methylation levels of the HBV and human genomes for the integrated and unintegrated alleles. Detailed results of the HBV integrants (PreC, Precore; C, Core; PreS, Presurface; S, Surface; X, X) and flanking host genomes (position, chromosome, location of the genome, and gene names) are shown. DNA methylation of the integrated HBV genome as well as the flanking human genome was examined by allele-specific DNA methylation analysis using bisulfite pyrosequencing. The HBV genome often showed significant methylation when integrated into highly methylated sites in the human genome; however, the HBV genome remained largely unmethylated when integrated into unmethylated regions. Methylation levels of orthologous loci in HepG2.2.15 cells and in PBLs of a healthy volunteer were examined and compared to the methylation levels at the same (empty) target sites of PLC/PRF/5 cells. Methylation levels of orthologous loci in HepG2.2.15 cells and PBLs were generally similar to those of PLC/PRF/5 cells. Similarly, methylation levels of orthologous loci in PLC/PRF/5 cells and in PBLs of a healthy volunteer were examined and compared to the methylation levels at the same (empty) target sites of HepG2.2.15 cells. Methylation levels of orthologous loci in PLC/PRF/5 cells and PBLs were generally similar to those of HepG2.2.15 cells. (×) The desired quantitative methylation levels were not obtained because of technical difficulties with the sequences that were being analyzed.











