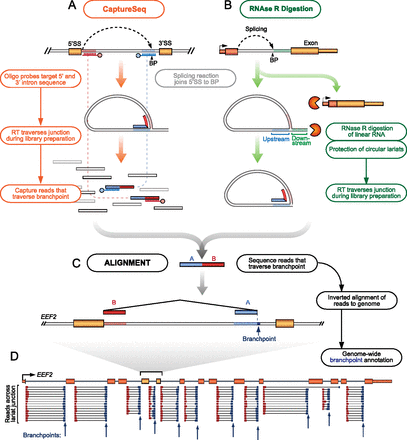
Identification of splicing branchpoints. Splicing joins the 5′ splice site to the branchpoint (BP) nucleotide near the 3′ end of the intron to form a lariat. Reverse transcriptase can traverse the branched 2′ to 5′ junction to generate informative sequencing reads (blue/red reads) that indicate the branchpoint location when aligned to the genome. We pursued two alternative strategies to enrich for sequenced reads containing branchpoints. (A) CaptureSeq (left orange pathway) uses labeled oligonucleotide probes to capture informative reads for targeted sequencing. (B) RNase R (right green pathway) digests linear mRNAs and selectively purifies circular RNAs including intron lariats. (C) Following sequencing, informative reads are split and inverted for alignment to the reference genome (black lower pathway), with 3′ termini of split alignment indicating the branchpoint nucleotide. (D) Example of informative alignments used to identify branchpoints (blue arrows) within the EEF2 gene.











