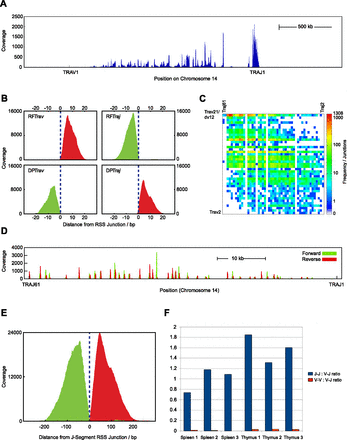
EC-seq enriches Tcra locus-associated circularized excision material. (A) Gross coverage of perfectly aligned read-pairs (AA) across the mouse Tcra locus on Chromosome 14 in whole thymus-captured EC-seq material. (B) Meta-analysis of Vα-Jα RF read-pairs (top) or DP read-pairs (bottom) in EC-seq libraries showing close association of reads with known Vα element subregion RSSs (left) or Jα element subregion RSSs (right). The data set comprises thymic and splenic EC-seq replicates. Overlay of forward reads is shown in green; overlay of reverse reads is shown in red. Broken vertical lines indicate the RSS cleavage site. (C) Heatmap analysis of 31,535 RSS-associated Vα-Jα RF junctions from EC-seq metadata displaying the diversity of recombined Vα-Jα ECs. Data are from three thymic and three splenic replicates. ECs are only shown for active elements with uniquely mappable sequences. (D) EC-seq pile-up of Jα-Jα RF read-pairs across the Jα segment subregion showing clustering of Jα element RSSs. Forward reads are shown in green; reverse reads are shown in red. (E) Meta-analysis of Jα-Jα subregion RFs relative to known Jα region RSS sites. Forward reads are shown in green; reverse reads are shown in red. (F) Ratio of Jα-Jα (blue bars) and Vα-Vα (red bars) to Vα-Jα RFs in three thymic and three splenic EC-seq libraries.











