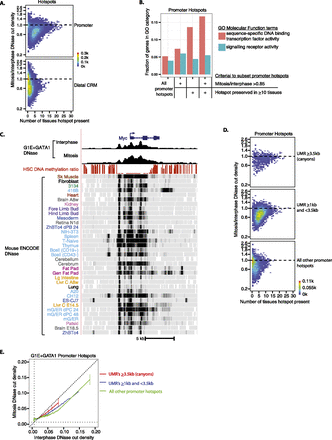
Promoters that are accessible across many murine tissues are marked by high mitotic accessibility and large DNA hypomethylation domains. (A) Promoter and distal CRM hotspots in G1E + GATA1 are shown in scatterplots (binned 2D density plot) of mitosis-to-interphase accessibility ratio versus the number of murine tissues in which the promoter hotspot overlaps at least one DNase hypersensitive site (DHS). Dashed horizontal line marks where mitotic and interphase DNase sensitivities are equal. (B) The fraction of promoter hotspots at genes encoding for sequence-specific transcription factor genes is specifically enriched by applying each of the two criteria (mitosis-to-interphase accessibility ratio and tissue preservation of hotspot) individually and together. (C) Myc is an example of a locus demarcated by a DNA methylation canyon. Shown are interphase and mitotic DNase accessibility profiles from G1E + GATA1, DNA methylation ratios in mouse HSCs, and DNase accessibility profiles from across Mouse ENCODE cell or tissue types, with dark bands representing DNase-sensitive regions. (D) G1E + GATA1 promoter hotspots are shown in scatterplots (binned 2D density plot) of mitosis-to-interphase accessibility ratio versus the number of murine cell or tissue (out of 45) DHS that is present. The panels are divided into promoter hotspots that overlap UMRs of the indicated size ranges. (E) Mitosis versus interphase DNase read densities of G1E + GATA1 promoter hotspots are shown as moving means color-coded by overlap with UMRs of the indicated size ranges, with error bars denoting SEM from biological replicates (n = 3). Error bars for the UMRs ≥1 kb and <3.5 kb (blue line) are omitted to avoid obscuring the difference between the red and green curves.











