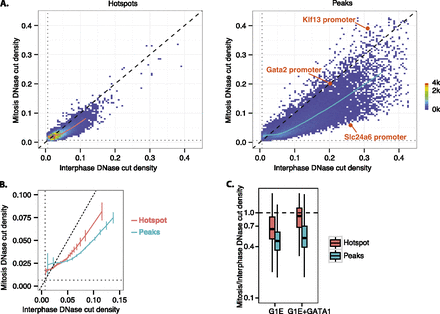
Chromatin accessibility is widely preserved during mitosis, with reductions occurring preferentially within narrow, hypersensitive DNase peaks. (A) After obtaining the union of the regions defined by hotspots/peaks across all samples, the mitotic versus interphase library size-normalized read densities were obtained from reads pooled from biological triplicates. Shown are mitotic versus interphase scatter plots (binned 2D density plots) for G1E + GATA1 cells. The color scale indicates the density of data points within each bin. The dashed diagonal line marks where mitotic and interphase read densities are equal. The overall trend is summarized by the moving mean (curve overlaid on plot) obtained from dividing the x-axis into bins consisting of ∼1000 hotspots or peaks. Gray dotted horizontal and vertical lines mark the estimated read density of inaccessible background regions, defined as all regions outside of hotspots. Data points corresponding to individual promoter peaks shown in Figure 2B are highlighted. The same graphs for G1E cells are shown in Supplemental Figure S6. (B) A zoomed-in view of the juxtaposition of moving means of hotspots and peaks in A. Error bars denote SEM of read densities from individual biological replicates (n = 3). (C) Box plot summaries of the mitosis-to-interphase ratio of the library size-normalized read densities for hotspots and peaks in G1E and G1E + GATA1, using reads pooled from biological triplicates. The horizontal dashed line marks where mitotic read density is equal to interphase read density.











