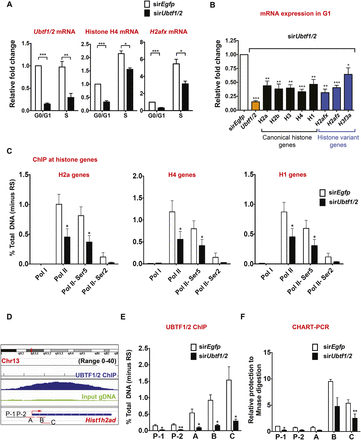
UBTF1/2 regulates histone gene expression by mediating Pol II recruitment. (A) NIH3T3 cells were transfected with sirEgfp or sirUbtf1/2#1 for 48 h, then collected and incubated with Vybrant DyeCycle Violet stain (Life Technologies). Cells were then analyzed using BD FACSAria, and G1 and S phase populations were sorted based on DNA content. Total RNA was extracted and Ubtf1/2, Histone H4, and H2afx mRNA levels were determined by reverse transcription qPCR. mRNA levels were normalized to beta-2-microglobulin (B2m) mRNA and expressed as fold change relative to sirEgfp/G0/G1 (n = 4). (B) Total RNA samples from G1 population of NIH3T3 cells transfected with sirEgfp or sirUbtf1/2#1 as in A were analyzed by reverse transcription qPCR for mRNA expression of various canonical and variant histone genes as indicated (n = 4). (C) UBTF1/2 mediates Pol II recruitment, initiation, and elongation at histone genes. qChIP analysis of the histone H2a, H4, and H1 genes in sirEgfp- or sirUbtf1/2#1-transfected NIH3T3 cells using antibodies against Pol I (n = 2), total Pol II (n = 7), phospho Pol II-Ser5 (n = 4), or phospho Pol II-Ser2 (n = 4). The percentage of DNA immunoprecipitated with the indicated antibodies or RS was calculated relative to the unprecipitated input control. Percentage of DNA of RS controls was subtracted. (D) Ubtf1/2 knockdown leads to increased DNA accessibility at Hist1h2ad. Screenshots of IGV with the mapped reads from UBTF1/2 ChIP and input gDNA in NIH3T3 to Hist1h2ad. Primers used for qPCR are indicated (Supplemental Table 21). (E) qChIP analysis of UBTF1/2 binding at Hist1h2ad in sirEgfp or sirUbtf1/2#1 transfected NIH3T3 cells. qChIPs were performed as described in Figure 4C (n = 3). (F) Chromatin remodeling of the Hist1h2ad by UBTF1/2. Nuclei from NIH3T3 cells transfected with either sirEgfp or sirUbtf1/2#1 for 48 h were incubated with or without MNase. Extracted gDNA was subjected to qPCR using primers outlined in D. MNase accessibility was expressed as a percentage of undigested gDNA samples (n = 3). In all graphs (A–F), error bars represent Ave ± SEM, (*) P-value < 0.05, (**) P-value < 0.01, (***) P-value < 0.001, compared to corresponding sirEgfp samples.











