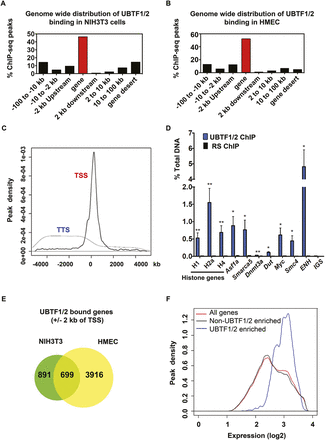
ChIP-seq analysis of UBTF1/2. Distribution of UBTF1/2 binding sites in NIH3T3 (A) and HMEC (B) with respect to RefSeq genes as determined by Sole-Search. (C) Average UBTF1/2 ChIP-seq enrichment profiles at all unique TSSs and TTSs in the mouse genome ± 5 kb. (D) qChIP analysis of UBTF1/2 binding in NIH3T3 cells to ChIP-seq genomic regions that reside within the indicated genes. The percentage of DNA immunoprecipitated with anti-UBTF1/2 or rabbit serum (RS) antibodies was calculated relative to the unprecipitated input control (n = 3–7; Ave ± SEM). (*) P-value < 0.05, (**) P-value < 0.01, compared to corresponding RS samples. Amplicons at the enhancer (ENH) and intergenic spacer (IGS) of rDNA were used as a positive and negative control for UBTF1/2 binding, respectively. Primers to core histone genes recognize multiple genes within each class (Supplemental Table 19). (E) Venn diagram indicating the overlap of UBTF1/2-bound genes between the NIH3T3 and HMEC cell lines. (F) Density histograms of gene expression levels in exponentially growing NIH3T3 cells for all genes, genes with significant UBTF1/2 ChIP-seq peaks < 2 kb from their TSS, or genes with no UBTF1/2 binding at their TSS. Statistical significance between groups was assessed using t-tests.











