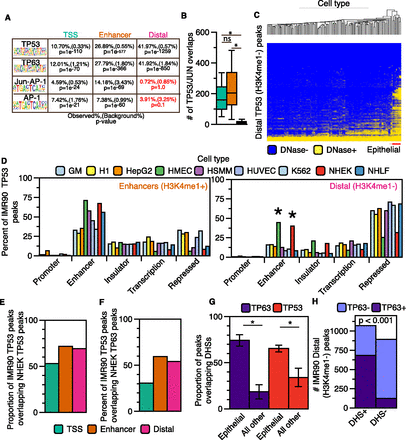
Distal (H3K4me1−) TP53 peaks bind to accessible chromatin in a cell-specific manner. (A) Transcription factor motif analysis for enhancer and distal TP53 peaks using HOMER. (B) Boxplot of JUN/FOS/TP53 colocalization at the three categories of TP53 binding sites. Asterisks denote that the P-value for the pairwise comparison is less than 0.05 (Wilcoxon rank sum test). (C) Hierarchal clustering (Euclidean distance) of the overlap between TP53 distal (H3K4me1−) peaks and DHSs from the ENCODE Project (minimum of 1-bp overlap between TP53 and DHSs; additional information in Supplemental Table S5). (D) Analysis of the overlap between TP53 enhancer (orange) and distal (pink) peaks with ChromHMM-defined genomic regulatory regions from nine human cell types. Similar functional ChromHMM categories were grouped into single categories (i.e., strong and weak enhancers grouped together). TP53 peaks were allowed to overlap multiple ChromHMM regulatory regions, and each region was reported (minimum of 1-bp overlap). (E) The proportion of IMR90 TP53 peaks overlapping TP53 binding sites from cisplatin-treated normal human epithelial keratinocytes (NHEK). (F) The proportion of IMR90 TP53 peaks overlapping TP63 binding sites from cisplatin-treated NHEK. (G) The proportion of TP63 (NHEK) and TP53 (IMR90) binding sites overlapping regions of DHSs from epithelial cell types and all other cell types with available data. P-values generated using a two-tailed t-test (P-values < 0.001). (H) Dependency of TP53 distal (H3K4me1−) peak overlap with DHSs (normal human epithelial keratinocytes) on co-occupancy with TP63: P-value < 0.001 (χ2 test).











