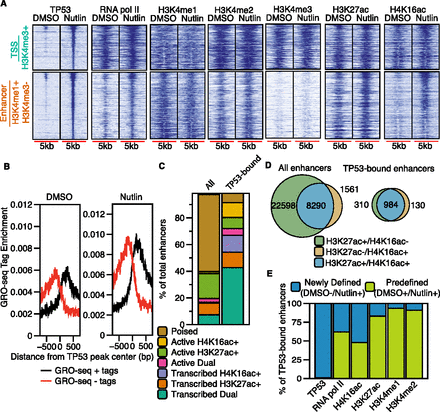
TP53 binds to established H3K4me1-marked enhancers and recruits RNA pol II and H4K16ac. (A) Heatmaps of TP53, RNA pol II, H3K4me3, H3K4me1, H3K27ac, and H4K16ac enrichment within a 5000-bp window (±2500 bp from the TP53 peak center) for TSS (H3K4me3+) and enhancer (H3K4me1+/H3K4me3−) TP53 peak types after DMSO or nutlin treatment. (B) Average GRO-seq profiles (normalized to 1 × 10−7 reads) at TP53 enhancer (H3K4me1+/H3K4me3−) peaks after DMSO or nutlin treatment. (C) Distribution of combinatorial histone modifications and RNA pol II enrichment at TP53-bound H3K4me1+ enhancers compared to the genome-wide complement of H3K4me1+ enhancers. (Poised enhancers) H3K4me1+; (active enhancers) H3K4me1+ and at least one acetylation event (H3K27ac or H4K16ac); (transcribed enhancers) H3K4me1+, RNA pol II+, and at least one acetylation event (H3K27ac or H4K16ac). Dual enhancers are occupied by both H4K16ac and H3K27ac. (D) Venn diagram representation of the overlap between H3K27ac+ and H4K16ac+ enhancers for all genome-wide and TP53-bound enhancers. (E) Analysis of the preestablishment of occupancy for each indicated factor at TP53-bound enhancers. Predefined enhancers are those in which the surveyed factor was significantly enriched (MACS-defined) during the DMSO treatment (no TP53 activation), whereas newly defined enhancers are those in which the enrichment of each factor was dependent on treatment with nutlin.











