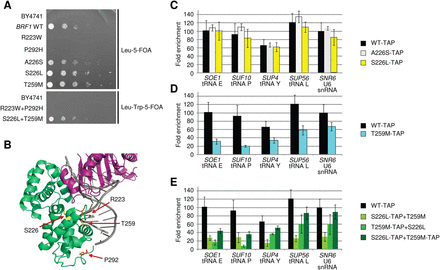
BRF1 mutations cause growth defects and reduced target promoter occupancy. (A) BRF1 mutations affect cell growth. Spot dilutions of the variants introduced into a BRF1 knockout strain grown at 30°C. Wild-type (WT) and variant Brf1 were encoded on plasmids. For the combination of two mutations, two plasmids were used, each harboring one mutation and a distinct marker. (5-FOA)5-fluoroorotic acid. (B) Three-dimensional modeling of human BRF1 missense alterations. The four identified amino acid substitutions were mapped to the structure of human TFIIB (green) in a complex with TBP (purple) and DNA (pdb code 1C9B) (Tsai and Sigler 2000). Amino acids affected by mutations in families 1 and 2 are shown in yellow and orange, respectively. (C–E) BRF1 variants show a decreased occupancy of tRNA promoters in yeast. Fold enrichments of ChIP experiments performed with tandem affinity purification (TAP)—tagged BRF1 variants in yeast. Data are presented as mean ± SD. (C) Mutations A226S to serine or S226L show no effect on fold enrichment compared to WT. (D) Mutation T259M leads to decreased fold enrichment on tRNA genes. (E) Combination of the two variants results in a much lower occupancy of the S226L and T259M variants. Summing up both signals results in less occupancy of the two mutated BRF1 than the WT BRF1.











