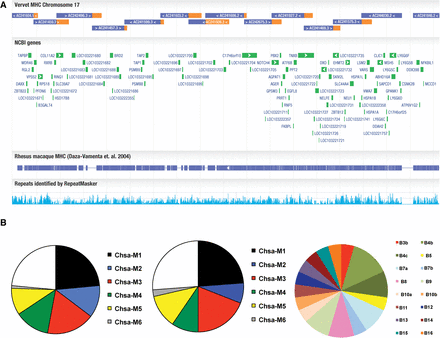
The characterization of the vervet MHC region and its diversity. (A) A tile path of sequenced and assembled BACs were aligned to a single haplotype region of the rhesus macaque MHC (Daza-Vamenta et al. 2004); vervet sequence aligned in the correct orientation with macaque is colored blue, and gaps are denoted as missing color. (B) Pie charts show MHC haplotype distribution in (left) captive, (middle) Caribbean, and (right) West African vervets. Six ancestral MHC class I haplotypes (B1–B6) identified by pyrosequencing accounted for all major haplotypes observed in vervets (n = 51) from US primate centers. The remaining haplotypes reflected simple recombination events between these six ancestral haplotypes. The distribution of MHC class I haplotypes of feral vervets (n = 21) from St. Kitts is virtually indistinguishable from that of US primate center monkeys. Eleven feral vervets from Ghana exhibited at least 16 distinct Chsa-B haplotypes. Only the Chsa-B5 haplotype (yellow slice) was identical with the class IB region of the B5 haplotype in St. Kitts-origin vervets. Chsa-A haplotypes in the individuals from Ghana also exhibited much greater diversity compared to the Caribbean-origin population (data not shown).











