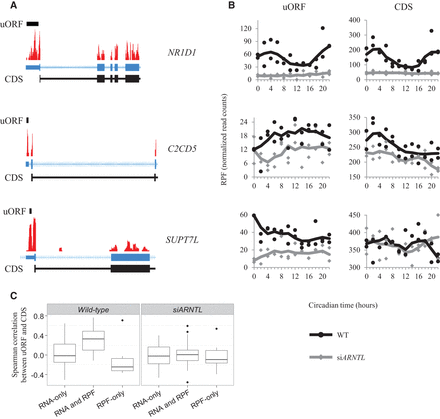
Translation of upstream open reading frames. (A) Coverage plots for RPF data mapped to the NR1D1, C2CD5, and SUPT7L loci are plotted. The locations of the uORFs are identified by the black bar above the coverage plot. The gene models for these genes are displayed in blue below the coverage plots. Blocks correspond to exons, and lines indicate introns. The thick blocks indicate the annotated coding region, while the thin blocks correspond to the annotated untranslated region. The chevrons within the intronic lines indicate the direction of transcription. CDS regions are displayed in black below the gene models. The regions displayed are truncated for visualization purposes. (B) RPF traces from the uORF and coding sequences are plotted as a function of time. The black traces are from the wild-type RPF libraries, while the gray traces are from the siARNTL RPF libraries. (C) Box plots for the distribution of Spearman correlation coefficients between uORF and CDS data are displayed for each of the three gene groups (RNA-only cyclers, RNA and RPF cyclers, RPF-only cyclers). These distributions are plotted for both the wild-type and siARNTL data.











