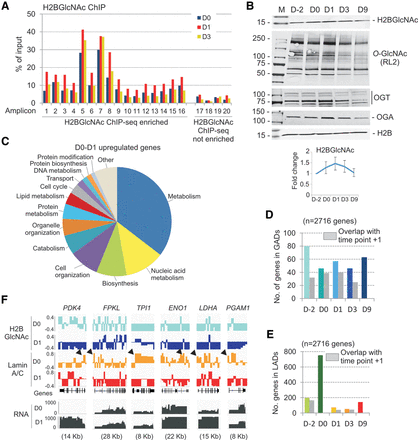
Adipogenic induction results in up-regulation of metabolic genes, lamin A/C dissociation from these genes, and in a transient increase in H2BGlcNAc. (A) ChIP-qPCR analysis of H2BGlcNAc on enriched and nonenriched loci on D0, D1, and D3 of differentiation. (B) Western blot analysis of H2BGlcNAc, total O-GlcNAc (RL2 antibody), OGT, OGA, and H2B throughout differentiation. Graph shows mean H2BGlcNAc levels at each time point relative to D−2 (levels standardized to H2B; mean ± SD of four experiments). (C) Gene ontology enrichment classification for the 2716 genes up-regulated on D1 (see Supplemental Fig. 4A,B). Note the high proportions of metabolic genes. (D,E) Localization of D0-D1 up-regulated genes in GADs (D) and lamin A/C LADs (E), and maintenance of these genes within GADs or LADs between consecutive differentiation time points (gray bars). (F) H2BGlcNAc and lamin A/C ChIP-seq profiles on PDK4 and indicated glycolytic HIF1A-target genes. Arrows point to losses of lamin A/C from promoters, coinciding with transcriptional up-regulation of the genes. RNA-seq profiles are shown (scale 0–1000 FPKM).











