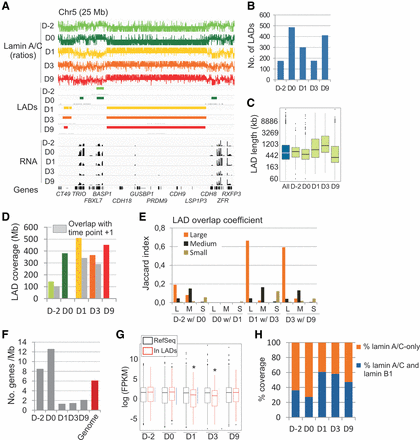
Lamin A/C LADs are reorganized by induction of adipogenic differentiation. (A) Lamin A/C ChIP-seq enrichment profiles showing ChIP/input ratios (scale −0.4 to +0.4 centered on 0), LADs mapped with EDD, and RNA-seq gene expression profiles (scale 0–1000 FPKM). (B) Number of LADs identified at each differentiation stage. (C) Median LAD lengths at all differentiation time points confounded (All) and at each time point. (D) Genome coverage by LADs (color bars) and conservation of LAD coverage between two consecutive time points (gray bars). (E) Coefficient of overlap of small, medium, and large LADs (Jaccard indices) with large (L), medium (M), and small (S) LADs between time points (x-axis). (F) Gene density within LADs and in the whole genome. (G) Median expression level of genes within LADs and of RefSeq genes: (*) P < 10−5, t-test with Bonferroni correction. (H) Proportion of lamin A/C LAD coverage at each time point, which overlaps with lamin B1 LADs mapped by DamID in fibroblasts (Guelen et al. 2008).











