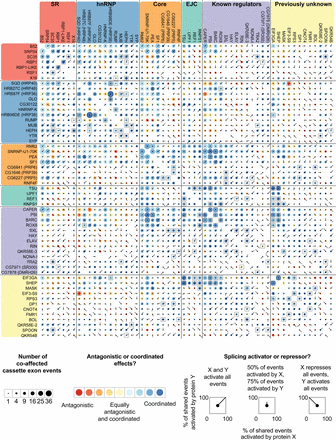
Antagonistic and coordinated effects of splicing regulators. Comparisons of cassette exons coaffected by each pair of proteins are represented as a matrix. Within each group (e.g., SR, hnRNP, Core, etc.), the proteins are ordered by the number of splicing events they affect, as shown in Figure 1. Information in the upper and lower portions of the matrix is identical. The size of each circle gives the relative number of coaffected events, and the color indicates if the pair acted antagonistically (one protein activates, the other represses) or coordinately (both proteins activate or both proteins repress). Overlaid spokes indicate what proportion of shared events is activated or repressed by each protein in the pair, thus giving more detailed information on the nature of the antagonistic or coordinated effects. Pairs of proteins that have a significant overlap in the number of coaffected events are indicated with black squares (Fisher's exact test, Bonferroni-corrected P < 0.05).











