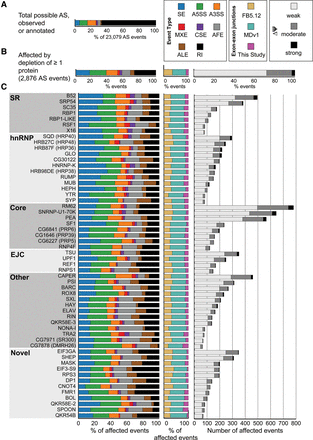
Alternative splicing (AS) events affected by depletion of 56 RNA binding proteins. (A) The proportion of each type of AS event that could potentially be regulated by each protein. (B) The observed proportion of each type of splicing event affected by depletion of at least one protein is shown alongside the type of exon–exon junctions involved in the splicing event. (C) The number, magnitude, type of AS, and type of exon–exon junction affected by each individual knockdown is shown. Each protein is categorized as SR, hnRNP, core splicing factor, exon junction complex (EJC), other known, or novel splicing regulator protein. The legend in the top right indicates the color codes used for the splicing event types. (SE) skipped/cassette exon, (A5SS) alternative 5′ splice site, (A3SS) alternative 3′ splice site, (MXE) mutually exclusive exons, (CSE) coordinate skipped/cassette exon, (AFE) alternative first exon, (ALE) alternative last exon, (RI) retained intron. In addition, the legend indicates whether the exon–exon junctions were found in FlyBase 5.12, MDv1 (Graveley et al. 2011), or novel to this study, as well as whether the magnitude of splicing changes observed were weak, moderate, or strong.











