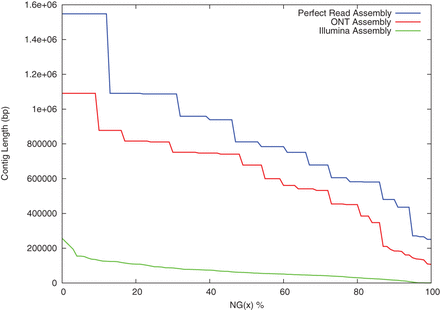
NG-graph of a simulated perfect read assembly, the corrected Oxford Nanopore assembly, and an Illumina-only assembly. The curve extends the common N50 metric to trace the contig size such that the top x% of the genome is assembled into contigs this size or larger. The Oxford Nanopore assembly is substantially more contiguous across the entire size spectrum and is far closer to the perfect read assembly than the Illumina-only assembly. Notably, the N50 contig length for the Oxford Nanopore-based assembly is 678 kbp compared to ∼60 kbp for the Illumina assembly and is quite close to the 811 kbp perfect read assembly N50.











