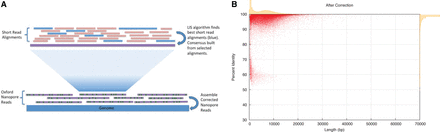
(A) Nanocorr workflow. Short high-identity reads are aligned to raw ONT reads. The best overlapping set is determined by the LIS algorithm, and a consensus sequence of these alignments is built using pbdagcon. Error-corrected reads can then be assembled using a long-read assembler. (B) Post-Nanocorr correction read length and accuracy. Scatter plot with marginal histograms summarizing the percent identity of reads after correction for W303. Average identity before correction is ∼68% for all iterations of flow cells, while the average post-correction identity was >97%.











