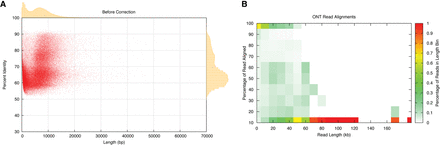
(A) Oxford Nanopore read lengths and accuracy. Scatter plot of read length versus accuracy with marginal histograms summarizing the raw ONT alignments. (B) Heatmap of Oxford Nanopore read lengths and accuracy. Each cell represents a summary of how reads of different lengths align. Each color represents the fraction of reads in a given read-length bin. Maximal alignment efficiency is observed between 10 and ∼40 kb, while fragments longer than 80 kb are virtually unalignable.











