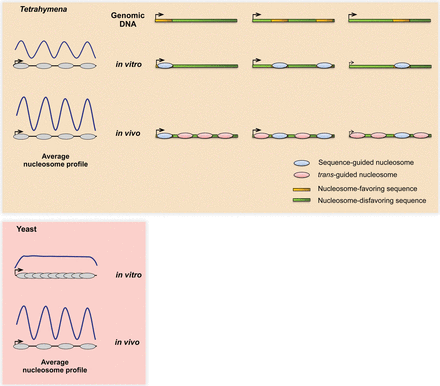
Contrasting mechanisms may underlie conserved nucleosome patterns in vivo between Tetrahymena and yeast. The Tetrahymena genome is GC-poor and is generally unfavorable for nucleosome formation. The majority of Tetrahymena genes encode nucleosome-favoring sequences at subsets of standard positions downstream from TSSs, which might in turn facilitate nucleosome positioning in and around these regions in vivo. On the other hand, yeast genes generally show no such DNA-guided specificity near TSSs, instead relying mainly on trans-acting factors to generate the distinctive nucleosome organization in vivo. As a result, the average in vitro and in vivo nucleosome patterns appear similar in Tetrahymena but not yeast.











