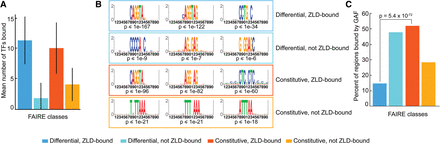
GAF likely functions with Zld to define chromatin accessibility in the early embryo. (A) Mean number of transcription factors (TFs) bound to each class of FAIRE peaks. Colors indicate the different FAIRE classes. Error bars, SD. (B) Position weight matrices for the motifs enriched in the FAIRE peaks for each class compared with all of the top 5000 FAIRE peaks identified in yw embryos. Hypergeometric P-values are shown below each motif. The top three motifs for each class are shown (for additional motifs, see Supplemental Table 1). (C) Percentage of FAIRE peaks in each class that overlap a GAF-binding site as identified by ChIP-seq in 0- to 8-h embryos (Negre et al. 2011). Hypergeometric P-value is shown.











