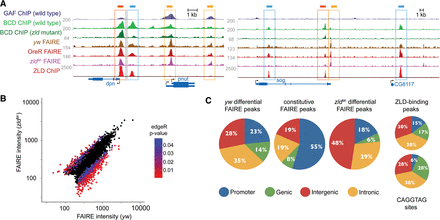
Zld is required to maintain or establish discrete regions of open chromatin. (A) Normalized FAIRE-seq and ChIP-seq read profiles as labeled on the left for two genomic regions. FAIRE data for yw and zldM- embryos (this study). FAIRE data for 2–4 h OreR (wild-type) embryos from McKay and Lieb (2013). ChIP data for Zld from Harrison et al. (2011), for Bcd from Xu et al. (2014), and for GAF from Negre et al. (2011). Genes are shown at the bottom with arrows to indicate direction of transcription. Boxes indicate different classes of FAIRE peaks: blue box, differential, Zld-bound; dark orange box, constitutive, Zld-bound; and light orange box, constitutive, not Zld-bound. (B) Scatter plot of the FAIRE signal from yw embryos versus the FAIRE signal from zldM- embryos. Black dots indicate the union set of regions identified by FAIRE in both yw and zldM- embryos. Colored dots indicate differential peaks identified by edgeR. Colors represent P-values (<0.05) as indicated by the scale. (C) Pie charts showing the genomic distribution of FAIRE peaks, Zld-binding sites, and CAGGTAG motifs to promoters, genes, introns, and intergenic regions as shown.











