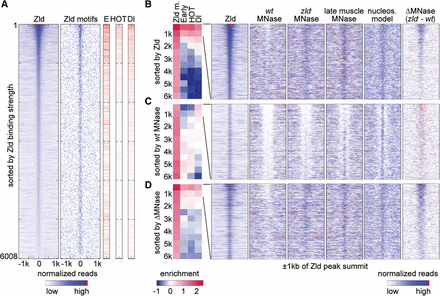
Zld binding and changes in nucleosome occupancy correlate with early enhancer activity. Analysis of 6008 Zld-bound regions that are >1 kb away from a TSS. In the heatmaps, normalized MNase-seq and ChIP-seq data for each region are aligned to the Zld summit, and 1-kb regions to each side are shown. (A) All 6008 non-TSS Zld-bound regions ranked by Zld summit reads from high to low show that Zld binding strongly correlates with Zld motifs and that the higher the Zld peak ranks, the more frequently early enhancers (E), HOT regions (HOT), and Dl peaks (Dl) overlap. The Zld peak rank also correlates with the degree of hotness, i.e., the number of TFs bound, and Dl binding strength, which are shown as degree of red in the same data. White indicates non-HOT regions. (B–D) The 6008 Zld-bound regions are ranked by Zld summit reads from high to low as in A (B); wt MNase (within 250 bp of Zld summits) from low to high (C), and ΔMNase (the read count difference between zld− and wt) from high to low (D). The ranked data were then divided into bins of 500 regions (except for the last bin, which has 508 regions), and enrichment values for each bin are shown (blue for depletion and red for enrichment). The enrichment of Zld motifs (Zld m.) was calculated over genome background, while the enrichment of early enhancers (Early), HOT regions (HOT), and Dl peaks (Dl) was calculated over the average of the 6008 Zld-bound regions. Note that the enrichment at the top two bins is strongest when ranked by Zld summit reads, still strong when ranked by ΔMNase, and the lowest when ranked by wt MNase. Heatmaps for the top two bins (1000 regions) in each ranking is shown to the right for the following data: Zld binding, wt MNase, zld− MNase, MNase profiles of 14–17 h muscle tissue (late muscle MNase), predicted nucleosome model (nucleos. model) (Xi et al. 2010), and ΔMNase. Note that the nucleosome occupancy in zld− embryos resembles that of late muscle tissue, where Zld is also absent, as well as the predicted model, suggesting that in the absence of Zld, nucleosome occupancy is governed by DNA sequence features.











