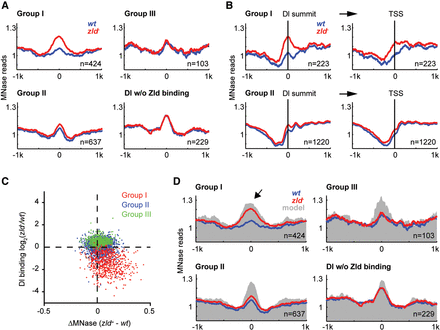
Loss of Dl binding in the absence of Zld is associated with increased nucleosome occupancy. Nucleosome occupancy was measured by MNase-seq (see Methods). (A) Metaprofiles (wt in blue, zld− in red) of Zld-bound Dl peaks that are >1 kb away from a TSS are shown for the three Dl-peak groups, as well as Dl peaks that do not colocalize with Zld binding as control. The normalized MNase reads were aligned at the Dl summit, and average reads within 1 kb distance are shown. (B) Metaprofiles of Zld-bound Dl peaks that are ≤1 kb away from a TSS are shown for Groups I and II. The normalized MNase reads were either aligned at Dl summits (left) or the nearby TSSs (right). Note that the increased nucleosome occupancy in zld− within Group I is much more pronounced on the left, arguing that the effect is directly due to Zld binding and not due to loss of transcription at these genes. (C) Scatter plot showing the correlation between ΔMNase (x-axis) and the fold change in Dl binding (y-axis) between zld− and wt embryos. Values were calculated using the reads within 125 or 250 bp of the Dl summit for Dl binding and MNase, respectively. Note the strong correlation for Group I Dl peaks (red). (D) Metaprofiles of the Dl-peak groups as in A, but with the addition of the average predicted nucleosome occupancy based on the underlying DNA sequence (gray) using a published prediction model (Xi et al. 2010). Note that the high and broad nucleosome occupancy of Group I regions in zld− is also predicted by the model (arrow), indicating that the role of Zld may be to tackle the intrinsically strong nucleosome barrier of 4–5 nucleosomes at these places, which would then help Dl access these regions.











