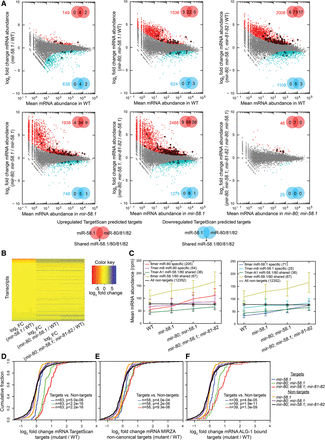
Mutations in individual miR-58 family members additively increase target RNA levels. (A) Plots of the log2 fold change in mRNA abundance between the conditions indicated on the y-axes against the expression level in the condition indicated on the x-axes. Transcripts deemed by DESeq (Anders and Huber 2010) significantly up-regulated or down-regulated transcripts with P(adjusted) < 0.01 are shown as red and blue dots, respectively, and their number is indicated in the upper right or lower right part of the plot. Significantly up-regulated (red) and down-regulated (blue) TargetScan-predicted miR-58.1, miR-80/81/82, and shared miR-58.1/80/81/82 targets are marked with bold dots, and their numbers are indicated in the Venn diagrams. (B) Heatmap indicating differential mRNA expression levels (log2 fold change [FC]) of 16,309 transcripts across different miR-58 family mutants. (C) Average mRNA abundances (in reads per million [rpm]) of groups of transcripts identified in all four conditions containing different seed matches—8mer, 7mer-m8, 7mer-A1, 6mer—and without seed match (in wild type, mir-58.1, mir-80; mir-58.1, and mir-80; mir-58.1; mir-81-82) indicate average contributions of miR-58 family members in their target up-regulation. TargetScan-predicted miR-58.1– and miR-80/81/82–specific and overlapping targets show similar additive trends. Error bars, SD. (D–F) Cumulative distributions of log2 fold changes of the following and 518 nontargets in different miR-58 family mutants compared with WT: (D) 63 TargetScan-predicted targets, (E) 58 noncanonical targets identified by MIRZA (score cutoff > 10), and (F) 39 ALG-1–bound miR-58 family targets (Grosswendt et al. 2014). P-values were calculated using a KS test comparing the fold change distributions (log2) of targets and nontargets.











