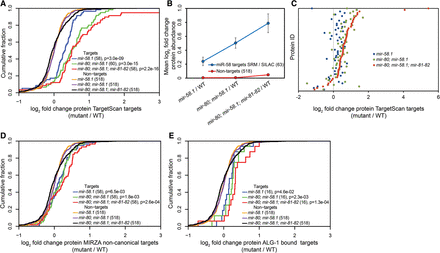
A combined selected reaction monitoring (SRM) and SILAC approach reveals additive target protein regulation of miR-58 family members. (A) Cumulative distributions of log2 fold expression changes of the 63 TargetScan-predicted miR-58 targets in the mir-58.1, mir-80; mir-58.1, and mir-80; mir-58.1; mir-81-82 mutants relative to the wild type (WT) indicate increased protein abundance with each miR-58 family mutation introduced. Quantified TargetScan-predicted targets identified in SRM, fractionated SILAC, and unfractionated SILAC were compared to a group of 518 nontargets identified in unfractionated SILAC measurements for which we also had transcript abundance data. P-values were calculated using Kolmogorov–Smirnov (KS) test comparing the fold change distributions (log2) of targets and nontargets. (B) Mean log2 fold changes of protein abundances of miRNA targets and nontargets in the different miR-58 family mutants relative to the WT indicate a cumulative effect of miRNA mutations. (C) Differential expression of 55 TargetScan-predicted miR-58 family targets quantified by SRM and unfractionated and fractionated SILAC in mir-58.1, mir-80; mir-58.1, and mir-80; mir-58.1; mir-81-82 relative to the WT. The protein ID indicates the specific protein quantified in different mutants relative to the WT. Proteins are sorted according to increasing up-regulation in the mir-80; mir-58.1; mir-81-82 quadruple mutant. It is apparent that each additional miRNA mutations further increases the expression of TargetScan-predicted targets compared with random controls. (D) Cumulative distributions of log2 protein-level fold changes of 58 noncanonical targets identified by MIRZA (score cutoff > 10) quantified by unfractionated and fractionated SILAC and of 518 nontargets described above in different miR-58 family mutant backgrounds. P-values were calculated using KS test comparing the fold change distributions (log2) of targets and nontargets. (E) Cumulative distributions of log2 protein-level fold changes of ALG-1–bound miR-58 family targets and of 518 nontargets described above in different miR-58 family mutant backgrounds compared with the WT. Requiring that a target reported by Grosswendt et al. (2014) is supported by at least five reads yielded 43 miR-58 family targets; of these, we obtained SILAC data for 16 targets. P-values were calculated using KS test comparing the fold change distributions (log2) of targets and nontargets.











