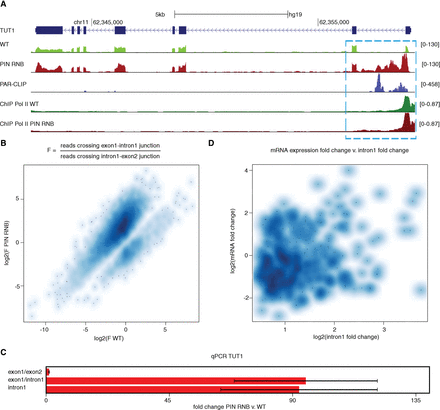
DIS3 degrades transcripts that arise from the premature termination of pre-mRNA transcripts. (A) A genome browser screenshot of the TUT1 gene, which is an example of the accumulation of prematurely terminated transcripts. The RNA-seq signal is supported by the PAR-CLIP and RNA polymerase II ChIP-seq signal. The mock control signals for Pol II ChIP, barely visible due to relatively low signal, are overlaid in scale on the respective tracks in gray color. (B) A higher number of fragments that overlap with the exon1/intron1 junction than the intron1/exon2 junction in DIS3 double mutant by RNA-seq. Transcripts that showed no change, defined here as the quotient between PIN RNB double mutants and WT, are not included in this plot. (C) Quantitative PCR validation of increases in levels of transcripts that cross the exon1/intron1 junction for the TUT1 gene. Bars represent the standard deviation for three biological replicates. (D) Lack of correlation between mRNA and intron 1 expression fold changes in DIS3 double mutants. The Spearman correlation coefficient = 0.0879, and the P-value = 0.0207. Only those mRNA transcripts that showed an increase in intron 1, but not in intron 2, were included in the analysis.











