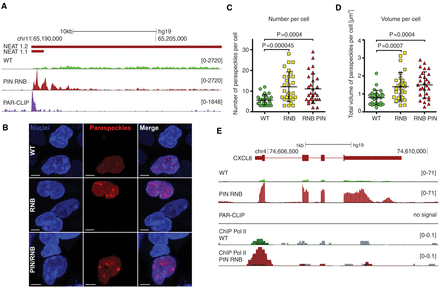
DIS3 dysfunction leads to NEAT1 lncRNA overexpression and the accumulation of paraspeckles. (A) A genome browser screenshot of the genomic region encompassing the NEAT1 locus with reads for mapped RNA-seq and DIS3 PAR-CLIP. (B) Fluorescent images of HEK293 cells transfected with constructs encoding the paraspeckle component NONO fused with mCherry. Nuclei were stained with Hoechst. White bars represent 5 µm. (C) The distribution of number of paraspeckles per cell in WT, RNB, and PIN RNB mutant cell populations. (D) Total volume of paraspeckles in a single cell, in WT, RNB, and PIN RNB mutant cell populations. (E) A genome browser screenshot of the genomic region encompassing the CXCL8 locus with mapped RNA-seq, RNA polymerase II ChIP-seq, and DIS3 PAR-CLIP reads (no signal). The mock control signals for Pol II ChIP are overlaid in scale on the respective tracks in gray color. Bars (C,D) represent the standard deviation from 28 measurements. P-values obtained using a t-test are shown. The uniquely mapped reads (A,E) are shown for the plus strand.











