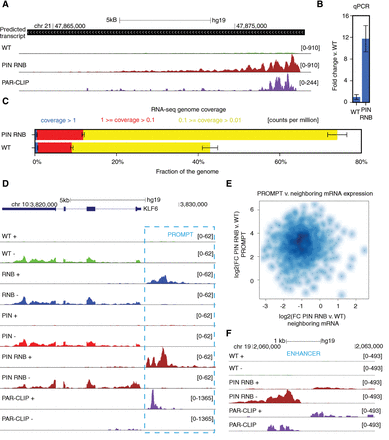
DIS3 inactivation leads to the accumulation of RNAs from unannotated parts of the genome and uncovers novel transcripts. (A) A genome browser screenshot of the genomic region encompassing one of the novel transcripts predicted by Cufflinks software with mapped RNA-seq reads for WT, PIN RNB double mutant, and DIS3 PAR-CLIP reads. The uniquely mapped reads are shown to indicate the minus strand. (B) Quantitative PCR validation of novel transcripts. Bars represent the standard deviation for three biological replicates. (C) The fraction of the genome covered by RNA-seq reads in WT and cell lines expressing the DIS3 PIN RNB double mutant. Coverage was measured by normalized read counts over each nucleotide. The sequencing depth for RNA-seq was ∼100 × 106 read pairs per sample. Bars represent the standard deviation for three biological replicates. (D) A genome browser screenshot of previously detected PROMPTs. RNA-seq reads mapped for WT, RNB, PIN, PIN RNB double mutants, and DIS3 PAR-CLIP. In D and F, uniquely mapped reads are visualized separately for the plus and minus strands. (E) The accumulation of PROMPTs does not correlate with the expression of neighboring genes. The relationship of expression fold changes between DIS3 WT and DIS3 PIN RNB double mutants for PROMPT and neighboring protein-coding transcripts, as shown on a logarithmic scale. (F) A genome browser screenshot of one of the enhancers accumulating eRNA in DIS3 double mutants. RNA-seq reads mapped for WT, PIN RNB double mutants, and DIS3 PAR-CLIP.











