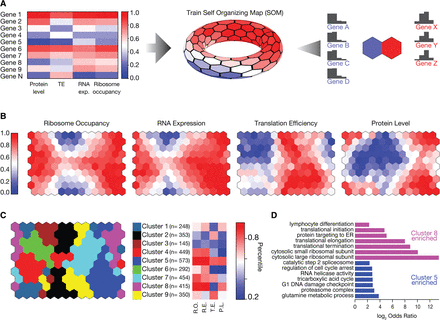
Ribosome occupancy correlates better with absolute protein levels than RNA expression and protein levels. (A) A self-organizing map (SOM) was trained using ribosome occupancy, RNA expression, translation efficiency (TE), and protein levels. These measurements were converted into their relative rank order before training. After training, each neuron in the SOM contains several genes sharing similar expression patterns. (B) Four different colorings of the trained SOM depict the mean ribosome occupancy, RNA expression, translation efficiency, or protein levels for each neuron. (C) Neurons of the SOM were grouped using affinity propagation clustering (Frey and Dueck 2007). Shared coloring between nodes indicates membership to the same cluster. For each cluster, the mean rank in ribosome occupancy (RO), RNA expression (RE), translation efficiency (TE), and protein level (PL) was shown for the representative neuron of the cluster. The number of genes in each cluster (n) is shown. (D) For four of nine clusters, significantly enriched gene ontology (GO) terms were identified (FuncAssociate; permutation-based corrected P-value < 0.05) (Supplemental Table S1; Berriz et al. 2009). For Clusters 5 and 8, selected GO categories were shown (log2 odds ratio). Supplemental Table S1 contains the full list of enriched terms.











