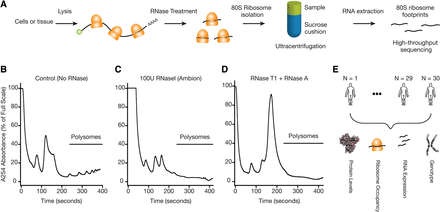
Choice of RNase is critical for generating ribosome profiling data. (A) A schematic representation of the ribosome profiling strategy is shown. A key step is the digestion of unprotected RNA segments with an RNase. The ribosome-protected RNA segments are isolated using a sucrose cushion and prepared for high-throughput sequencing. (B) Human lymphoblastoid cells (GM12878) were lysed in the presence of cycloheximide. The samples were ultracentrifuged through a 10%–50% sucrose gradient. Samples were fractionated while continuously monitoring absorbance at 254 nm. A representative polysome profile is shown. (C) Samples were prepared for ultracentrifugation as in B with the following exception: The cleared lysate was incubated with 100 units of RNase I (Ambion) for 30 min at RT before the ultracentrifugation. (D) Samples were prepared as in B, except 300 units of RNase T1 (Fermentas) and 500 ng of RNase A (Ambion) were used for the RNase digestion step. A complete digestion of polysomes into monosomes was observed. (E) Schematic representation of the data sets used in the current study. Genotype, ribosome profiling, RNA-seq, and mass spectrometry-based proteomics data were collected from lymphoblastoid cells derived from a diverse group of 30 individuals.











