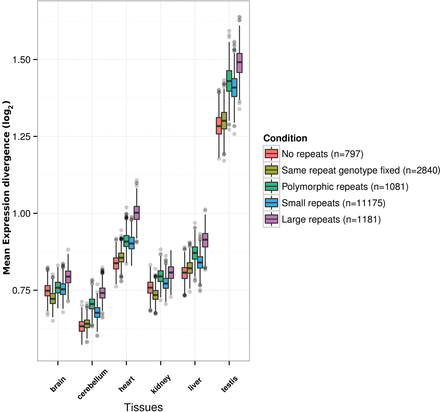
Relationship between expression divergence and within-species repeat genotype conservation in human and chimpanzees. Boxplots produced by resampling 1000 data points, each corresponding to the average log2-transformed expression divergence value between human and chimpanzee for a particular tissue and for genes associated to a particular category. The tissues are shown on the x-axis, and the y-axis corresponds to the absolute mean expression divergence between humans and chimpanzees. The categories considered are as follows: genes with no repeats in promoters (red), genes containing exclusively repeats in promoters that have the same repeat length fixed across all human and chimpanzee samples (olive), genes containing exclusively repeats that are polymorphic in human and chimpanzees (green), genes containing small repeats (repeat unit length of 1–5 bp and <100 bp total repeat length) in their promoter (blue), and genes containing large repeats (repeat unit length of 2–50 bp) in the promoter (magenta). Genes lacking repeats in promoters or repeats for which the same repeat genotype length is found across human and chimpanzee samples show the least amount of expression divergence for all tissues.











