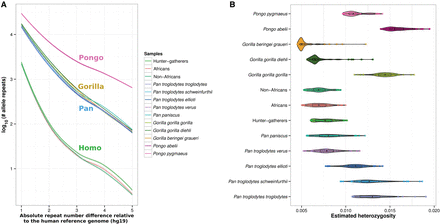
Repeat copy-number differences relative to human reference genome (hg19) and heterozygosity estimates for several nonhuman great apes species and human groups. (A) Absolute repeat number differences relative to the human reference genome estimated to occur for different human groups and subspecies/species of nonhuman primate taxa. The x-axis shows the number of repeat copy-number differences; the y-axis, the number of events for each repeat number difference in log10 scale. As expected, humans show the fewest differences relative to the human reference genome and are followed by chimpanzees, bonobos, gorillas, and orangutans. Within humans, African hunter-gatherers show the most variation, followed by Africans and non-Africans. (B) Heterozygosity estimates for different human groups and subspecies/species of nonhuman primate taxa. These results show great concordance with previous genetic surveys using millions of SNPs.











