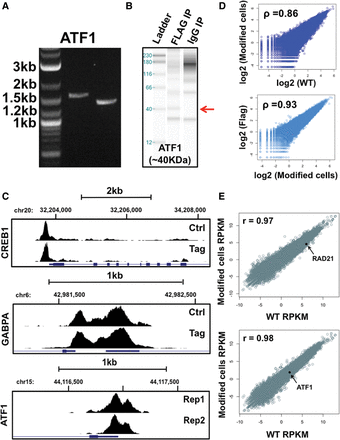
HepG2 ChIP-seq reproducibility and gene expression analysis. (A) PCR validation of ATF1 homologous recombination. From left to right, the gel image displays ladder, 5′ end, and 3′ end homology PCR products. Ladder band sizes are given at the left. Correct homologous recombination generates 1626-bp (5′ end) and 1433-bp (3′ end) gel bands. (B) IP Western blot validation of epitope-tagged ATF1. An ∼40-kDa protein band (marked by red arrow) corresponding to the predicted size of ATF1 is visible in IP Western blots using a Flag antibody but is absent in a control IgG pulldown. (C) DNA-binding protein read enrichment tracks on the UCSC Genome Browser are shown at distinct genetic loci. Data are given for both CETCh-seq (Tag, lower panels) and standard ChIP-seq using transcription factor antibodies (Ctrl, upper panels). The transcription factor name is displayed at the left of each image. For ATF1, technical CETCh-seq replicates are displayed (Rep1 and Rep2). (D) RAD21 rank correlations of normalized sequence read counts between CRISPR-modified HepG2 cells (Modified cells) and wild-type HepG2 cells (WT), both using a RAD21 antibody (top). (Bottom) RAD21 rank correlations of normalized sequence read counts between CRISPR-modified HepG2 cells using a RAD21 TF antibody (Modified cells) and CETCh-seq results of tagged RAD21 using Flag antibodies (Flag). Average rank correlations for all Flag and RAD21 replicate pairwise comparisons are given in the top left corner of each plot. (E) RNA-seq gene RPKM comparisons between CRISPR-modified HepG2 cells (Modified cells RPKM) and wild-type HepG2 cells (WT RPKM) are plotted for RAD21 (top) and ATF1 (bottom) experiments. Rank correlations and the location of tagged transcription factor RPKM values on each graph are displayed.











