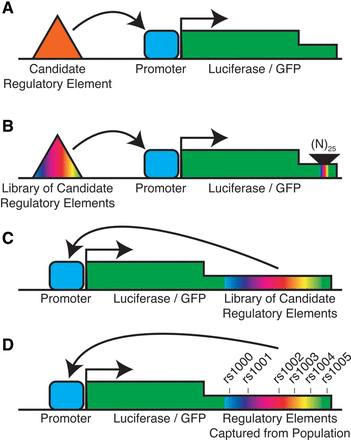
High-throughput reporter assays. (A) In a standard reporter assay, a candidate regulatory element is placed upstream of a reporter gene that is expressed from a constitutively active promoter. (B) In a high-throughput version of the same system, a random DNA sequence known as a molecular barcode is inserted into the 3′ UTR of the reporter gene, and a library of candidate regulatory elements are placed upstream of the promoter. Each individual candidate regulatory element is physically linked to a unique molecular barcode. Measuring the expression of each molecular barcode can then be used to estimate the activity of the associated regulatory element. (C) An alternative strategy is to clone the library of candidate regulatory elements directly into the 3′ UTR of the reporter gene. By that construction, the regulatory element controls its own expression, which can be measured with paired-end high-throughput sequencing. (D) The preceding strategy can be modified for genetic studies by cloning genetically diverse regulatory elements captured from donor genomes into the reporter gene. By that construction, each allele is expressed at a level that is directly related to its regulatory activity.











