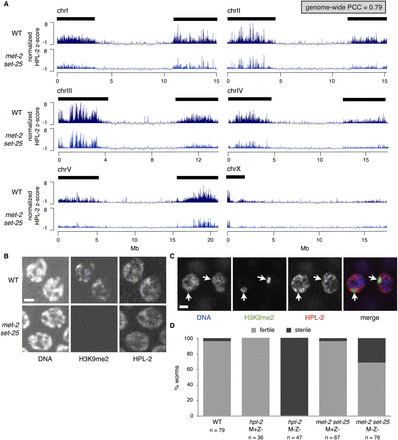
HPL-2 associates with chromatin independently of H3K9me. (A) Normalized mean Z-scores for HPL-2 ChIP-chip signal over the length of each chromosome in wild-type (WT) embryos and met-2 set-25 double mutant embryos, which lack H3K9me (Supplemental Fig. S5). The genome-wide PCC of HPL-2 ChIP-chip signal between WT and met-2 set-25 is 0.79. Chromosome arms are marked with black bars (see Fig. 1). Supplemental Figure S6 shows differently normalized HPL-2 ChIP-chip signal. (B) Immunofluorescence images of DNA, H3K9me2, and HPL-2 in WT and met-2 set-25 hermaphrodite germline nuclei. WT and met-2 set-25 samples were stained in parallel, and images were acquired using identical settings. Scale bar, 2 μm. (C) Immunofluorescence images of DNA (blue), H3K9me2 (green), and HPL-2 (red) in two WT male (XO) germline nuclei. Arrows point to single unpaired X chromosomes. Scale bar, 2 μm. (D) Percentage of hermaphrodites that were fertile (gray) or sterile (black) at 25°C in wild type (WT), hpl-2 M+Z-, hpl-2 M-Z-, met-2 set-25 M+Z-, and met-2 set-25 M-Z-. (M) Maternal supply of gene product, (Z) zygotic synthesis of gene product.











