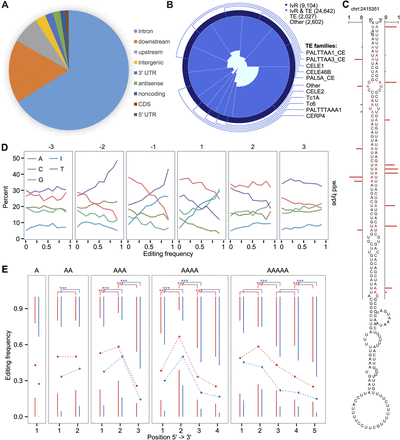
Characteristics and sequence preference around the RNA editing sites. (A) Location distribution of RNA editing sites. (B) Overlap between the RNA editing sites and each of the three categories: inverted repeats that coincide with transposons (InvR & TE, royal blue), inverted repeats outside of transposons (InvR, midnight blue), transposon regions not containing inverted repeats (TE, light cyan). Indicated in parentheses are the numbers of overlapped sites, which are also represented by the colored areas. (C) A predicted stem–loop structure of an RNA from an editing locus (Chr I: 2415351). Edited adenosines are marked red, and the red bars denote editing frequency. (D) Nucleotide preference within 3 nt around the editing sites in the wild type. The RNA editing sites were divided into 10 groups according to their editing frequencies. (E) Editing frequency at different positions of adenosine homopolymers in the wild type (blue) and the adr-1 (red) mutant.











