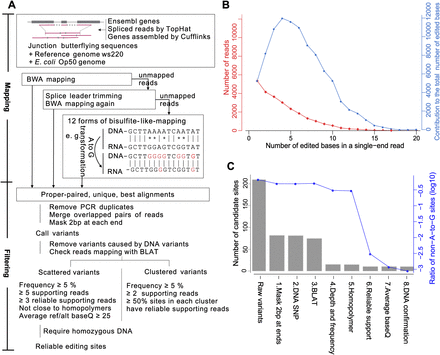
The computational pipeline for identifying RNA editing sites in C. elegans using whole-genome DNA-seq and strand-specific RNA-seq data. (A) Overview of the computational pipeline. This pipeline consists of three mapping steps. In particular, the ultra-edited reads are mapped using a bisulfite-seq-mapping-like approach. Multiple filters are applied to control false positives caused by library construction, sequencing, and misalignment. (B) The performance of the bisulfite-seq-mapping-like approach. The red curve shows the number of reads containing the indicated numbers of edited bases, and the blue curve shows their contribution to the total number of edited bases. (C) The performance of each filter in reducing the false positives.











