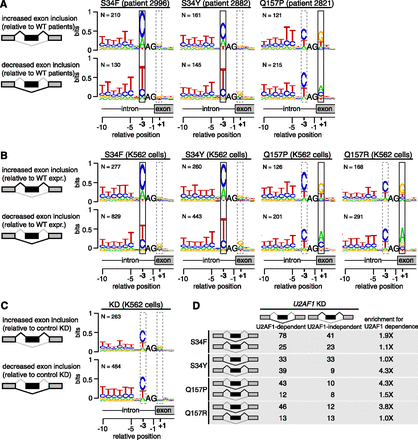
U2AF1 mutations alter 3′ splice site consensus sequences. (A) Consensus 3′ splice sites of cassette exons with increased or decreased inclusion in U2AF1 mutant relative to WT AML transcriptomes. Boxes highlight sequence preferences at the −3 and +1 positions that differ from the normal 3′ splice site consensus. (Vertical axis) Information content in bits; (N) number of cassette exons with increased or decreased inclusion in each sample. Data for all U2AF1 mutant samples is shown in Supplemental Figure S6. (B) As in A, but for K562 cells expressing the indicated mutation versus WT. (C) As in A, but for K562 cells following U2AF1 KD or control KD. (D) Overlap between cassette exons that are promoted or repressed by mutant versus WT expression (rows) and U2AF1 versus control KD (columns) in K562 cells. The third column indicates the enrichment for U2AF1 dependence, defined as the overlap between exons affected by mutant U2AF1 expression and exons repressed versus promoted by U2AF1 KD.











